A protocol for applying low-coverage whole-genome sequencing data
STAR Protocols is an open access, peer-reviewed journal from Cell Press. We offer structured, transparent, accessible, and repeatable step-by-step experimental and computational protocols from all areas of life, health, earth and physical sciences.

Best practices for variant calling in clinical sequencing, Genome Medicine

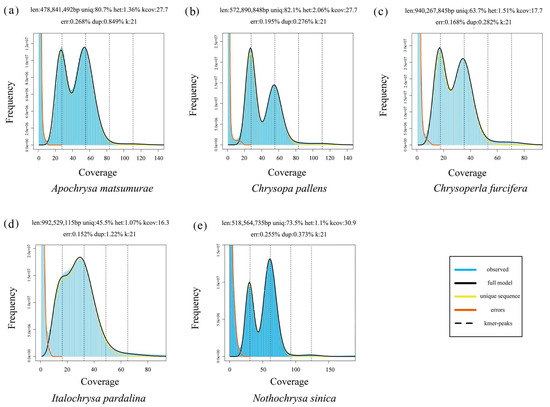
PDF) Analysis of population structure and genetic diversity in low-variance Saimaa ringed seals using low-coverage whole-genome sequence data

PDF) A protocol for applying low-coverage whole-genome sequencing data in structural variation studies

PDF) Analysis of population structure and genetic diversity in low-variance Saimaa ringed seals using low-coverage whole-genome sequence data

PDF) Analysis of population structure and genetic diversity in low-variance Saimaa ringed seals using low-coverage whole-genome sequence data

Copy number profiling using nanopore low-pass whole genome sequencing.
Biology, Free Full-Text

A benchmarking study of SARS-CoV-2 whole-genome sequencing protocols using COVID-19 patient samples - Abstract - Europe PMC
PDF) Mapping structural variations in and reveals vector–pathogen adaptation

Home - HumPOG

9149 PDFs Review articles in RHIPICEPHALUS

A) In this study we performed paired-end high coverage whole genome
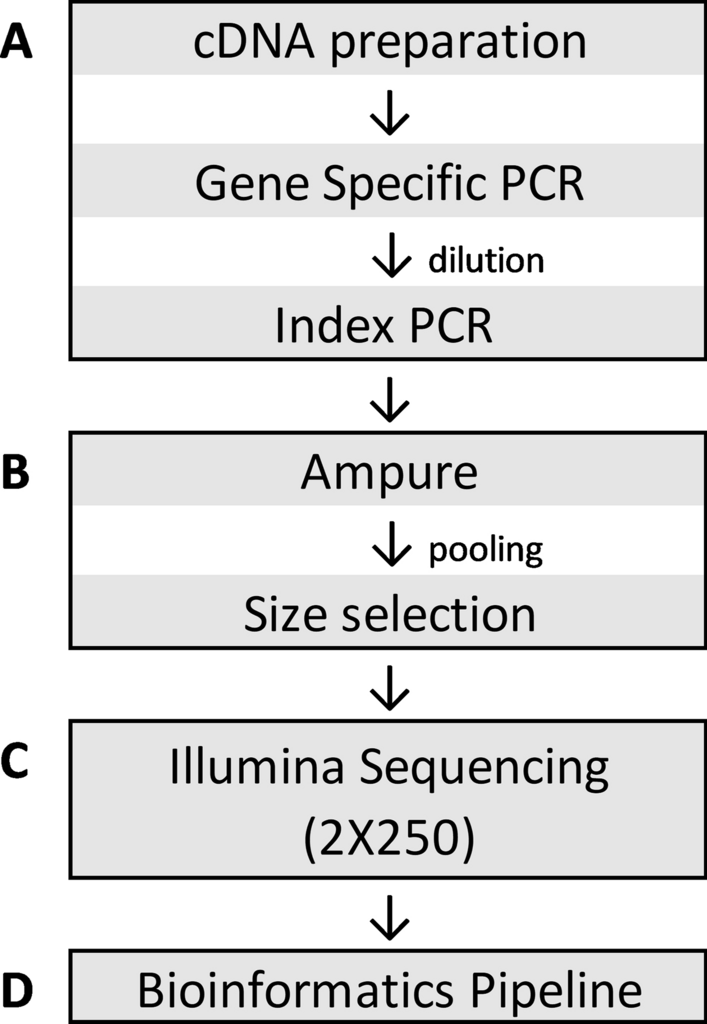
Development and validation of a high throughput SARS-CoV-2 whole genome sequencing workflow in a clinical laboratory

Bo XIE, Chinese Academy of Sciences, Beijing, CAS, Shanghai Institutes for Biological Sciences