Technical spotlight: Detecting small- and medium-length copy number variants by whole-genome sequencing
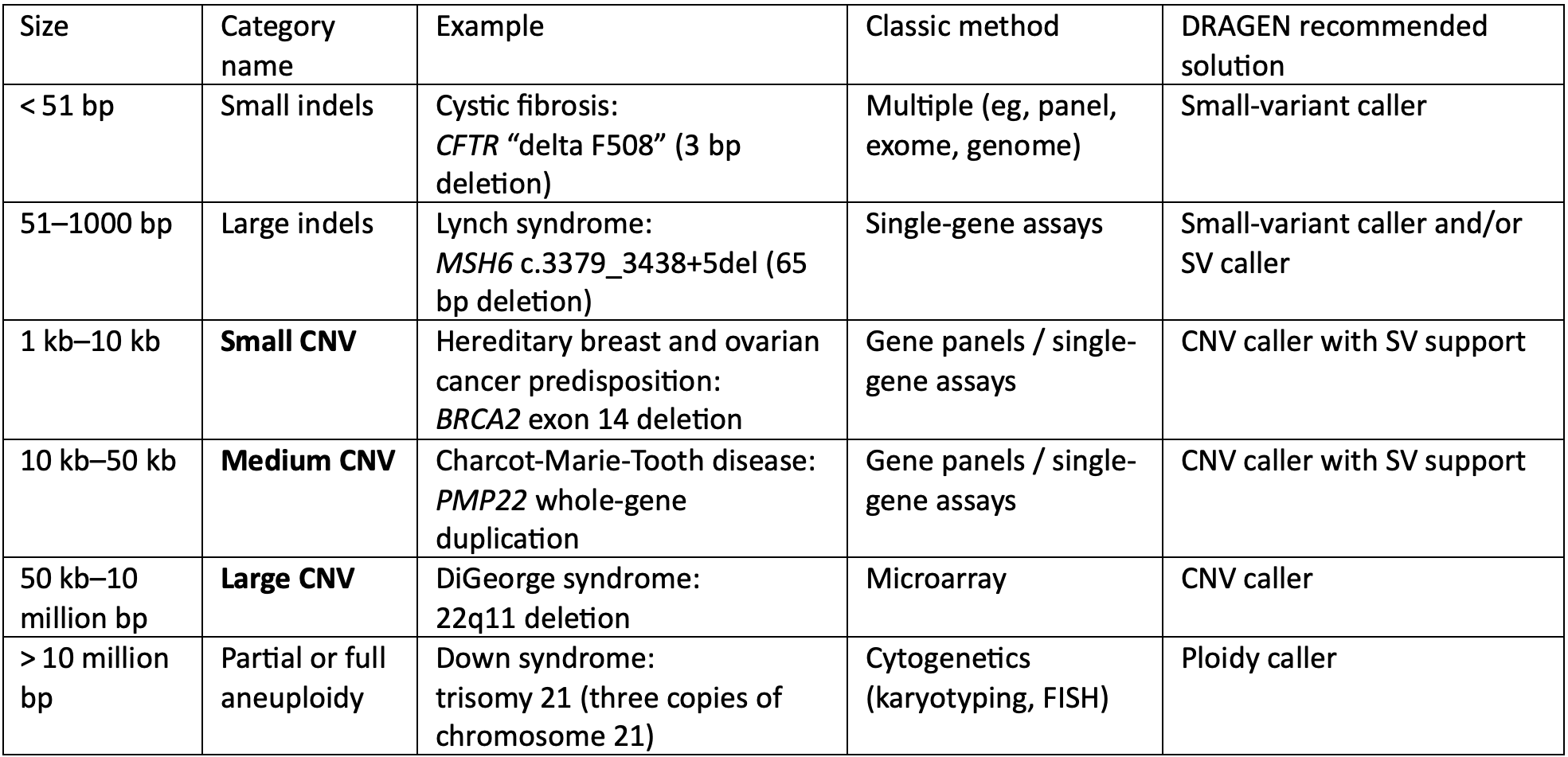
Historically, detecting different sizes of genetic variants has required using multiple different tests. By combining Illumina WGS with secondary analysis algorithms built into the DRAGEN Bio-IT Platform, researchers can achieve high-sensitivity detection of all these different variant types using a mixture of methods described here.

ClinSV: clinical grade structural and copy number variant detection from whole genome sequencing data, Genome Medicine
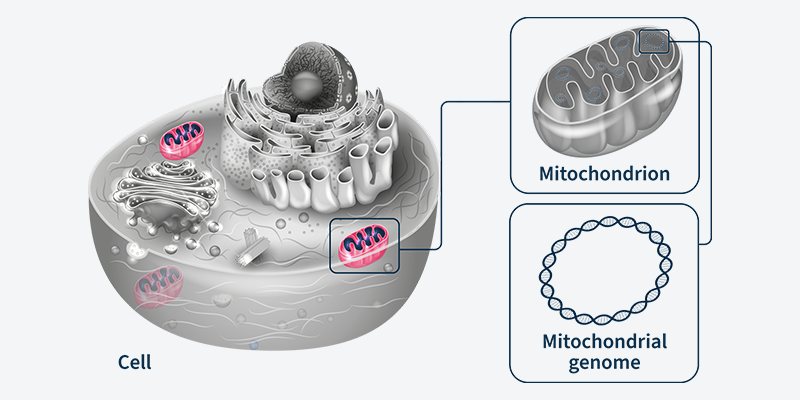
Continuing to crack the mitochondrial genetic code - SOPHiA GENETICS

Rami Mehio on LinkedIn: #emedgene #dragen #illumina

Detecting copy number variation in next generation sequencing data from diagnostic gene panels, BMC Medical Genomics
Copy Number Variation (CNV) Analysis

Samuel Strom, PhD FACMG on LinkedIn: whoa. I mean I knew thing

High-throughput sequencing technologies in the detection of livestock pathogens, diagnosis, and zoonotic surveillance - Computational and Structural Biotechnology Journal
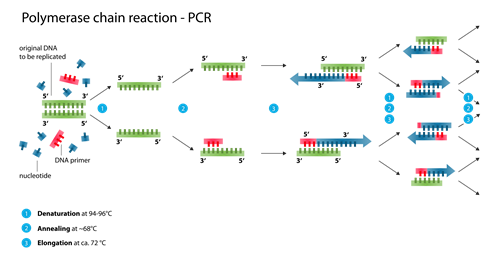
Amplicon Sequencing

Rami Mehio on LinkedIn: Using whole-genome sequencing to evaluate

Rami Mehio on LinkedIn: Illumina Acquires Edico Genome to
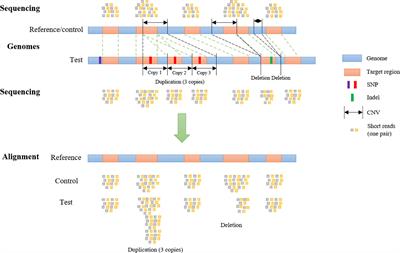
Frontiers SECNVs: A Simulator of Copy Number Variants and Whole-Exome Sequences From Reference Genomes
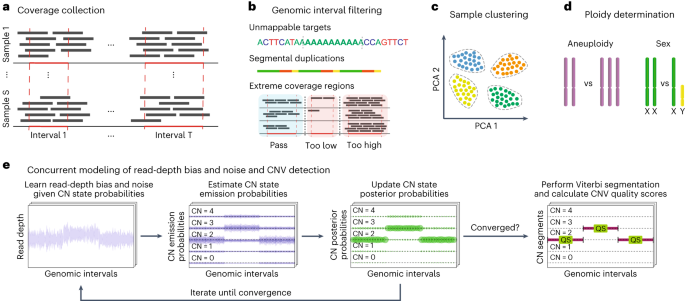
GATK-gCNV enables the discovery of rare copy number variants from exome sequencing data

Copy Number Variant Detection Using Next-Generation Sequencing - ScienceDirect

Genomics Articles Recent genomics discoveries by Illumina scientists

Copy Number Variation (CNV) Analysis